Make’ing Your Dependencies Explicit
Posted on 19 July 2017
Make’ing Your Dependencies Explicit
By Blair Archibald, University of Glasgow.
Building reproducible research workflows can be a messy business: data comes from many sources, it may need formatting, combined with other data and analysed in some way. Luckily, there is a whole host of software tools available to help manage some of this complexity (and hopefully let you keep your sanity!). In particular, (GNU) Make is ideally suited for the purposes of producing reproducible workflows. To see why let's join the FAKE research group.
The FAKE Research Group
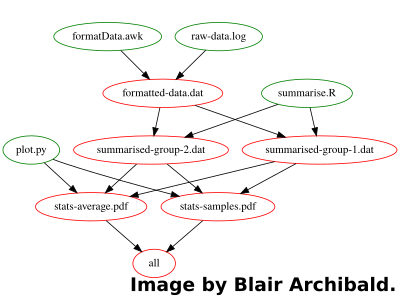
Welcome to FAKE, a data driven research group that makes heavy use of computational science to perform analysis for publication. Our first task is to get up to date with the current publication. Luckily our predecessor has left detailed written instructions of the data analysis workflow:
-
Run the formatData.awk script over the raw data to generate a tabular formatted output for later analysis
-
Use the summarise.R R script on the formatted data to create two new data sets: summarised-group-1 and summarised-group-2
-
You can then use the plot.py Python script on the two groups to generate the plots for the paper: stats-average.pdf and stats-sampled.pdf
These instructions contain lots of information: the scripts we require, the input and outputs that we expect, and the order the commands should be run in.
While these instructions are detailed enough for us to generate the required plots, what happens when new data appears or one of our scripts changes? We need to manually run each step again. We might say that this workflow is replicable but not automated.
A common method for automating this sort of workflow would be to write a script that glues together these commands. A popular scripting language for this purpose is Bash, giving us the following script:
#!/bin/bash awk -f formatData.awk raw-data.log > formatted-data.dat Rscript summarise.R formatted-data.dat python plot.py summarised-group-1.dat summarised-group-2.datWe can now re-run this script to handle any data or script changes, avoiding the need to go through each step manually. We now have an automated workflow.
But there's a problem, as we moved from the written instructions to the Bash script we lost information about the workflow! Importantly, the written instructions made it clear where the data sets summarised-group-1 and summarised-group-2 come from. Likewise, the Bash script does not document what is finally created by the Python script. What we really need is a tool that lets us both detail the commands to be run while making the input and output dependencies explicit. Enter Make.
Make
Make is a tool often at the heart of many software build systems (even new systems such as Automake and CMake tend to generate Make compatible files). Its popularity makes it available on a large number of platforms.
The core of Make is a Domain Specific Language that the user writes in a Makefile. The language specifies how to build the particular components (targets), from their dependencies [1]. Together we call the target/dependencies/command a rule. For example:
target : dependencies command ...When the user asks Make to build a particular target (by invoking make target), Make figures out the dependency graph and executes all the commands required to build that target.
This target/dependency/command structure of Make allows it to be used for much more than just software build systems. At its heart, the language simply provides a way to explicitly specify the input and output dependencies of a command/set of commands. In fact, we can think of it as follows:
outputs : inputs command ...The ability to explicitly specify input/output dependencies allows Make to be hugely powerful when creating replicable workflows.
Let's reimplement our data analysis steps using Make. Firstly, we format the data:
formatted-data.dat : formatData.awk raw-data.log awk -f formatData.awk raw-data.log > formatted-data.datThis says: “I know how to build formatted-data.dat if I have the formatData.awk script and the raw-data.log file, and I do that by calling awk on it and redirecting the output”. We have successfully encoded everything that was specified in step 1 of the workflow description.
What about step two? Here we use a lesser known feature of Make: it's possible to have any number of outputs (target names) per rule.
summarised-group-1.dat summarised-group-2.dat : summarise.R formatted-data.dat Rscript summarise.R formatted-data.datThe command is exactly what we had in the Bash script, but now we have explicitly stated that the command produces two new files. Again, no information is lost from the initial description.
Creating the plots follows the same pattern giving us the complete Makefile:
.PHONY : all # Build all pdf’s required for the paper all : stats-average.pdf stats-samples.pdf # Parse and format raw experiment data (Step 1) formatted-data.dat : formatData.awk raw-data.log awk -f formatData.awk raw-data.log >formatted-data.dat # Run statistical analyses (Step 2) summarised-group-1.dat summarised-group-2.dat : summarise.R formatted-data.dat Rscript summarise.R formatted-data.dat # Draw statistical plots (Step 3) stats-average.pdf stats-samples.pdf : plot.py summarised-group-1.dat summarised-group-2.dat python plot.py summarised-group-1.dat summarised-group-2.datAs calling Make with no target name, by default executes the first target in the file, to take advantage of this we create a PHONY target at the top of the file (PHONY essentially says it doesn't produce anything) To run the workflow we simply call "make" in the folder with the data/scripts, and it will figure out the correct command ordering automatically.
Comments are introduced using the # character allowing complicated targets to be further documented for future reference. Lastly, to make it easier for a user that doesn’t know which files the Makefile can produce, we provide a simple help target:
# Display the helpstring and list of targets .PHONY : help help : # “@” stops the Make from printing the command before execution @echo “FAKE Research Group Paper 1 Makefile” @echo “Targets:” @echo “all - build all targets” @echo “formatted-data.dat - parsed raw data file” @echo “summarised-group-1.dat summarised-group-2.dat - statistical analysis files” @echo “stats-average.pdf stats-samples.pdf - statistical plot files”This allows a user to type make help and receive a list of target names.
Unfortunately this can be tricky to manage as the Makefile increases in size. An alternative approach is to generate the help string from target comments as described in the Make Software Carpentry Lesson.
Benefits of Explicit Dependencies
Moving to Make and having explicit dependencies not only stops us losing workflow information but also get us a whole host of benefits for free:
-
Avoiding excessive recomputation: Make is smart enough to figure out that if no input dependencies for an output have changed, then it doesn't need to recompute the output at all, as it must be up to date. If we ran Make twice on the example above (without touching any files), only the first run would execute any commands.
Likewise, if we changed the summarise.R script the data formatting awk script wouldn't run again as Make knows the formatted data is already up to date. -
Running partial analyses is easy: Let's say we are doing a quick analysis and don't actually care about the plots. In the Bash version we need to edit the script to comment out the plotting step. With Make we just ask for the file we want, e.g., make summarised-group-1.dat, and it will only run the first two steps.
-
Parallelism: Since Make has the full file dependency graph it can figure out when two rules can be run in parallel. Using make -jN tells Make that it run N jobs simultaneously (if it can), where N should be roughly the number of cores you want to use.
Conclusion
We have seen how Make can be an effective tool for creating replicable research workflows. The key feature that makes this possible the ability to explicitly state the input and output dependencies of a command. This input/output information provides not only improved documentation for replicable workflows but allows further benefits such as avoiding wasteful computation, partial workflows and parallelism.
For your next workflow: Make your dependencies explicit!
Footnotes
[1] From the make website.

