Landscape archaeology and reproducible research at the 2017 Berlin Summer School
Landscape archaeology and reproducible research at the 2017 Berlin Summer School
Posted on 9 August 2017
Landscape archaeology and reproducible research at the 2017 Berlin Summer School
 By Ben Marwick, University of Washington and University of Wollongong.
By Ben Marwick, University of Washington and University of Wollongong.
Recently we concluded the 2017 Summer School on Reproducible Research in Landscape Archaeology at the Freie Universität Berlin (17th–21th July), funded and jointly organised by Exc264 Topoi, CRC1266, and ISAAKiel. With a group of 15 archaeologists and geographers from Berlin, Kiel and Cologne, we spent a week learning advanced geostatistics and how to make our research more reproducible.
Reproducibility is a relatively new concept for archaeology; it has only recently received some attention, and so we encountered many challenges in teaching and learning this unfamiliar subject. The general idea is that a piece of research, such as a publication, should contain enough information for another researcher to reproduce the results in that paper. In former times, when archaeology was simpler than today, this was relatively straightforward. However, that is not the case nowadays, with computers and complex software and algorithms playing a key role in our data analysis. Thanks to computers, the complexity of modern archaeology means that it is difficult to fit all the details of an analysis into the text of a typical journal article. There just isn't the space. This is a serious problem because the lack of detail in modern publications makes it hard for us to decide if the research is reliable or not and slows us down in reusing published research in our own projects.
In our Summer School, we explored solutions to this problem of reproducibility. Our focus was R, which allows us to write instructions for every step of our data analysis. Many archaeologists are now turning away from Excel and SPSS towards R to write scripts for for their data analysis. There are many advantages to this. R works on any computer and is free to install and use, unlike commercial products. It also has over 10,000 add-on packages, so there is tremendous flexibility in the types of analysis it can do. And, of course, because we write scripts to use with R, we have a record of our analysis that we can share with our publications. This means that people reading our publications can easily see all the decisions we made during our data analysis. They can take our code and change it to explore the effects of making different decisions in the analysis.
Our group were already advanced R users, so the challenge we faced was how to organise our projects in a simple, logical way to enhance reproducibility. We wanted to take advantage of existing conventions in R to organise our work in a way that makes it easy for other users to navigate. Our goal was to come up with some simple steps for getting organised with a new or ongoing project that uses R. We were also keen to investigate how we can use modern software engineering tools to automate some of the processes to check the reproducibility of our research. We wanted our final product to embody the best practices that we've read in several recent high-profile publications.

Our solution was one of the main results of our workshop. This is the new R package rrtools, or 'reproducible research tools'. This free and open source package provides instructions, templates, and functions for making a basic compendium suitable for reproducible research and writing a journal article or thesis with R. After much experimentation and trial and error in our group, we boiled the setup process down to eight quick and easy steps. These steps include:
1. Creating a new empty R package as the research compendium. This is important because most R users recognise the file structure of an R package. So they will know where to look for the different parts of the compendium.
2. Adding licenses to specify how we want the contents of the compendium to be reused by other researchers.
3. Connecting our compendium to GitHub, a web site for sharing code and data files. GitHub is also an excellent system for collaborating on writing papers and code and sharing files.
4. Adding some basic, structured machine-readable and human-readable metadata, instructions on how to cite the compendium, and instructions for potential contributors.
5. Adding a file structure and document templates to organise the typical components of a research project and accompanying journal article or book. For example, separate directories for data and code.
6. Adding a Dockerfile that contains a recipe for making an isolated computational environment for our analysis. This means we're not constrained to a specific computer with a specific setup to ensure that our analysis can be done correctly. We can create a Docker container to reproduce the exact computational environment that our original analysis was based on.
7. Connecting our repository to an online service that automatically tests our code each time we update it. This saves us from having to spend a lot of time testing our code—the Travis service runs the tests for us.
8. Adding tests for our custom functions. In case our works includes complex R functions, we should add tests to ensure that our function do what we expect.
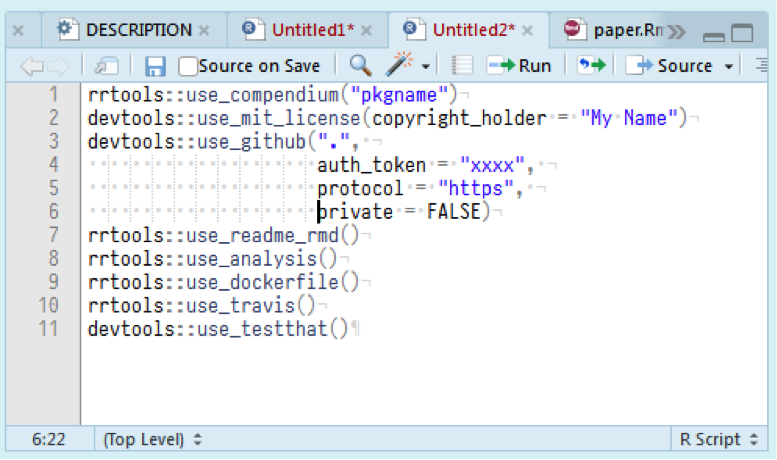
For each of these steps, we have a one-line function to complete the task. In less that five minutes, you can start from nothing, run these functions, and then have a complete research compendium. In our group, we mostly used RStudio to run R, but also some Emacs users. The eight functions described above save a lot of intermediate struggles with organising our work to be transparent, open and reproducible. The file structure generated by our package includes several template files. With these templates, we can start writing and doing data analysis right away.
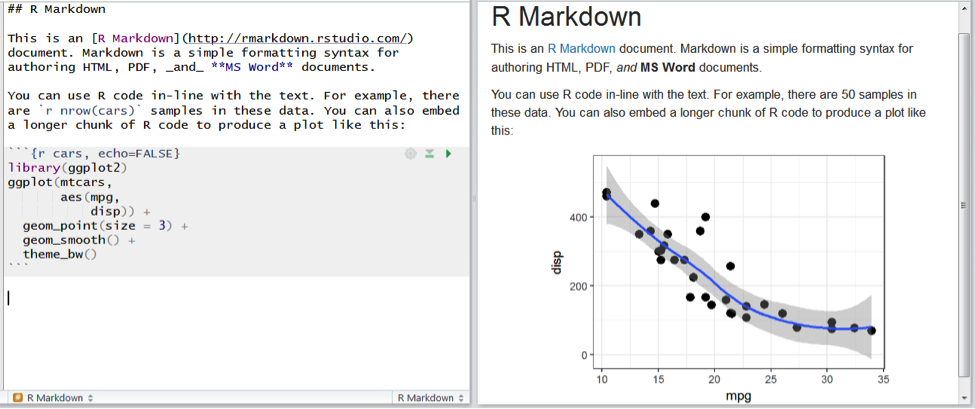
The writing system we use in this approach is called R Markdown. It's different from writing in a word processor because the document does not look exactly like how it will print out. Instead we see plain text, with some code-like formatting to specify bold, italics, etc. Markdown allows us all the usual scholarly writing tools, such as citing references (we especially recommend Zotero to help with this), adding tables and figures, and including numbered captions and cross-references. Using R Markdown, we can weave blocks of R code in between the paragraphs of text. These blocks of code can make plots or tables that will appear in the final document. It is possible to write entire journal manuscripts in this R Markdown system, as some of our group have already done.
We're very pleased with this outcome from our Summer School. This rrtools package was born off an intense effort by our group to find simple and easy methods to improve the reproducibility of archaeological research. We're already using it among our own projects. We invite anyone interested to take a look, try it out, and give their feedback.
