RightField - Stealthy semantic annotation with spreadsheets
Posted on 2 November 2021
RightField - Stealthy semantic annotation with spreadsheets
 By Stuart Owen and Katy Wolstencroft.
By Stuart Owen and Katy Wolstencroft.
This blog post has been written as an introduction to Rightfield ahead of the panel discussion: Do we have the right tools for research (5 Nov 2021, 2pm GMT). The panel is part of the Research Software Camp: Beyond the Spreadsheet.
Introduction
RightField is an open-source application that was designed to make collecting standardised, semantically annotated and machine-readable metadata simple and straightforward. It provides a mechanism for embedding ontology annotation into Excel spreadsheets. Researchers are not required to have detailed ontological and metadata knowledge, but they can benefit from better standardisation of data, whilst continuing to work with Excel. In addition, RightField automatically extracts metadata into RDF, meaning that data can be shared as interoperable Linked Data.
RightField was developed for the Systems Biology community to share Minimum Information Model-compliant data in the FAIRDOMHub (e.g. MIAME (Minimum Information About a Microarray Experiment). It therefore reduces the barrier to adopting metadata standards and their associated ontology terms for annotation.
Origin
Back in 2010, as part of the SysMO (Systems Biology of Microorganisms) project, we were involved with helping experimentalists improve their data management techniques. During this process we spent a lot of time with our users learning about how they collect and annotate data and the nature of the data they produced.
Data labels and values were often defined as plain text, which meant no consistency between different data sets and data producers, and no control or validation over what labels meant, which was prone to errors. The users knew about the use of ontologies and controlled vocabularies to annotate data - but this would be something they would do after the fact and found it cumbersome and complex.
We knew that good data management and collection requires well described data, and felt that some barriers would be removed if we could encourage standardised annotation at the time the data was created. We also wanted to encourage the use of ontologies and the semantic web, but keep it as straight-forward as possible, and hide away some of the complex terminology that can be discouraging.
Notably, we learnt that users were predominately making use of spreadsheets and they were by far the most common exchange format used. Spreadsheets provided a set of benefits including:
- They can be used offline, which was important at the time for being able to collect data directly within the lab.
- Data coming off the instruments in the lab could either be easily exported as Excel spreadsheets, or transferred easily from an intermediate format such as CSV or TSV.
- They are a self-contained package that can be shared easily through email, Dropbox, etc.
- They have a lot of features built in, and other tooling already available built around them.
- They were already a very familiar tool. Users were not required to learn something new and many already had advanced skills in their use.
When we started the requirements gathering process, we did so with the expectation of building a new tool or web interface to support data collection needs. However, it quite quickly became apparent that we needed to build our solution around the use of spreadsheets. Any attempt to impose a new tool would have been met with resistance, and trying to do so would have been wasted effort that would for the most part be ignored. We also wanted to avoid reinventing the wheel.
This led us to the concept of RightField. It would be a tool that would only be used by a domain specialist for generating well formed Excel spreadsheet templates. The domain specialists would be familiar with metadata management and ontologies and would therefore define which ontologies and which ranges of ontology terms were required for a given template. The experimentalists themselves would not see RightField, they would only see the spreadsheet templates, which were regular excel sheets with dropdown lists of terms to select from for specific fields. No additional tools were imposed upon them and they could continue to work as they normally would.
Design Ideas
When designing RightField we arrived at some key decisions:
No user should need to see RightField. They would work exclusively with spreadsheets, following the same process and workflow as they did before. The exception is the domain specialist that uses RightField to build the spreadsheet templates with integrated ontologies.
It would be a tool that generates spreadsheets, rather than a plugin that needs to be added to a spreadsheet. We wanted the barrier to be as low as possible, and interfere with the workflow of our users as little as possible. We wanted users to be able to use the spreadsheets generated directly without the additional step of installing a plugin. For similar reasons we wanted to avoid using VB scripts entirely. This leads on to the next point.
The spreadsheets should be compatible with Open Source alternatives that are compatible with the Excel file format, in particular LibreOffice (known at the time as OpenOffice). Although the Excel format was being used ubiquitously, many of our users used Linux as their primary operating system, and we didn’t want to force a change. The spreadsheets generated needed to be “vanilla” and simple enough to be interchangeable.
RightField itself should be open source, under a liberal license. We wanted the tool to be transparent in what it does and how it works, and free to be reused and repurposed without restriction. We were also encouraging the sharing of open data and open science, so we should do as we preach. We chose the BSD 3-clause license.
It should be easy to get the annotations out again. Either directly within RightField, and also in a way that can be programmatically integrated into another service.
Users of the spreadsheets should need no prior knowledge of the semantic web. We wanted to enable standardised, semantic annotation, without requiring users to become ontology experts.
It should be domain independent. Although its initial target was for experimentalists in Systems Biology, we recognised it would be useful in any domain that makes use of ontologies for annotation, so wanted to make sure the software avoided any domain specific references.
How RightField works
RightField is implemented in Java, which we chose due to our familiarity and also the actively maintained 3rd party libraries available to process and write Excel files (Apache POI) and process OWL ontology documents (OWLAPI).
When RightField is started, you can start by either loading an existing spreadsheet or starting afresh with an empty one. The spreadsheet is shown in a view similar to Excel, shown as individual sheets tied to a tab. There is some basic editing available, such as adding and removing sheets, copy and paste and delete. It would have been infeasible, and unnecessary, to recreate all edit and formatting features of Excel as the spreadsheet can be formatted in Excel before or after applying the ontologies in RightField.
Ontologies can be imported either from a file, via a URL, or from the BioPortal biomedical ontology repository. Once loaded, the ontologies will appear as a tree in a panel on the right, and multiple ontologies can be loaded simultaneously.
Individual spreadsheet cells, or a range of cells, can be selected for the addition of ontology features applied from the tree. These can be ontology classes or instances, and by default everything below the root node will be applied to the cells - although individual terms can also be removed if necessary. Optionally, a property can also be applied to the cell, which indicates something additional about the purpose. A cell can also be marked as being free text, and have a property applied to say what this is, such as a title or description from Dublin Core.
Once finished, the spreadsheet is exported and can be used as a regular Excel sheet. Cells that have had a list of terms applied will appear as a typical dropdown box, containing a flat list of the term labels. Details about the ontologies and the individual terms are held in “very hidden” sheets. The sheets contain standard data validations that produce the dropdown boxes, along with a corresponding column that holds the term URI, and additional information about the ontology, the root term, and the source of the ontology.
It is common to then do further edits to the spreadsheet after the export, such as adding formatting. Since it is vanilla Excel based on data validations, it is even possible to copy, paste and move around the marked up cells without any unwanted side effects.
The spreadsheet can be shared, and duplicated, to be filled out and completed by end-user scientists, then it can be reloaded into RightField and exported as basic RDF, or as CSV, that contains information about the values selected or filled out. This can also be achieved through a command line option, allowing it to be integrated into other software (as it currently is with FAIRDOM-SEEK). When RightField-enabled spreadsheets are uploaded to the FAIRDOM-SEEK, the semantic annotations contained within the sheet are extracted and used to automatically populate the metadata, which is subsequently stored as RDF.
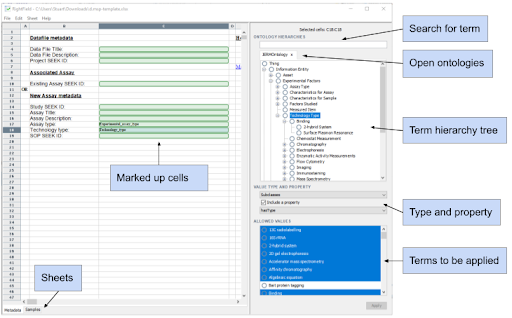
You can find a more detailed walk through, with additional screen shots in the RightField User Guide.
RightField on the Web
As previously mentioned, when gathering requirements from our users, one of our original constraints was that the solution should be available to use off-line. This was because an internet connection wasn’t always available at the point the data was collected and annotated. This led us to avoid a solution that was web based. However, times move on and we are increasingly asked about an online solution that can be built into data pipelines.
Fortunately, a nice side effect of our original red-line to stick to simple vanilla Excel, with no use of plugins or scripts, has provided an additional benefit. Since there is nothing special about the spreadsheets generated, they work with existing online tools with little additional work. The spreadsheets can be shared with Dropbox, Google Drive or OneDrive. They can be cloned, shared and collaborated on using Google Sheets or Office365 with no adverse side effects, and then exported again as Excel files which can be passed through RightField to extract the annotations.
Conclusion and future work
Rightfield has already been used to develop ontology-annotated, Minimum Information metadata templates for domains outside of systems biology. With each expansion into new domains comes more specialist requirements and also new ideas for new functionality or ways of making the application more sustainable. The Netherlands Bioimaging network (NL-BI) has adopted RightField for sharing standardised bioimaging data, specifically for high content screening and for zebrafish microscopy experiments. Through this preliminary work, we identified a number of functional augmentations that are required for bioimaging data, and also to make RightField easier to use for the NL-BI community.
In the NL-BI roadmap initiative at the Leiden Institute of Advanced Computer Science, we are working on enabling groups of spreadsheet cells to be annotated and referred to together. This will be achieved by introducing named graphs, allowing interconnections in the data to be more easily described, and allowing dynamic changes to subsequent ontology term lists based on earlier choices. These augmentations would also greatly benefit the FAIRDOM-SEEK user-base, allowing all annotations to be automatically extracted and directly stored as RDF; and increase the usage of RightField as a stand-alone application throughout the life sciences
